A team of McGill researchers published a paper in Nature Methods showcasing neuromaps, an open-source Python toolbox that allows neuroscientists to analyze brain imaging data using a consistent set of tools and compare it with a curated brain-map database. PhD candidate Justine Hansen, one of the paper’s first authors, spoke with the The McGill Tribune about how neuromaps works and what she hopes it will accomplish.
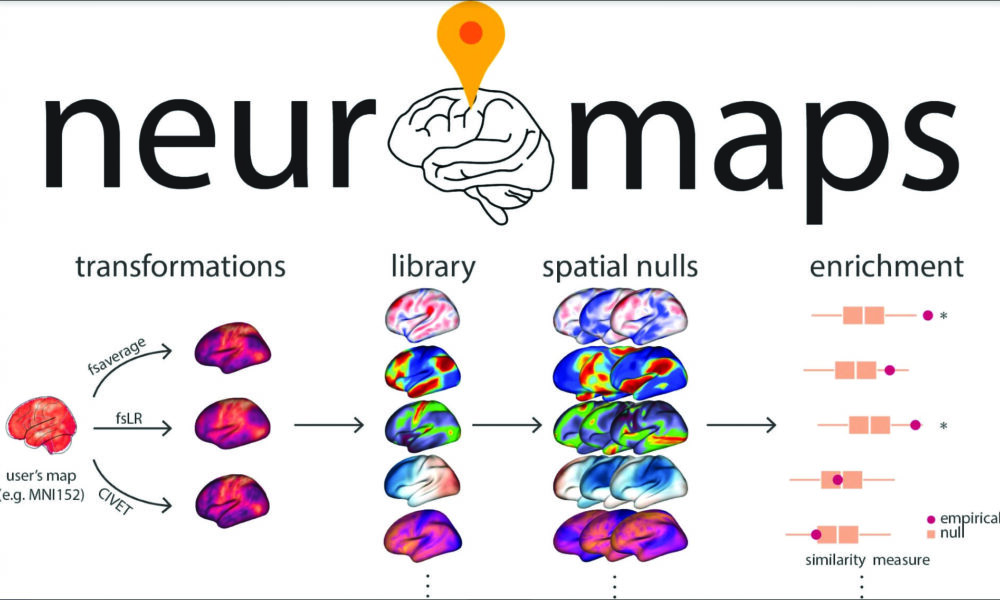
Neuromaps gives researchers a set of software tools to transform data from one format to another without having to perform the calculations themselves. At the same time, it pulls together a database of about 40 standard brain maps that researchers can compare their data to, and a set of “spatial nulls,” which are tests for statistical significance in the data. Combined, these features make the process of analyzing, comparing, and drawing conclusions from data more streamlined and reliable.
At its core, neuromaps aims to simplify brain imaging data, as the data is incredibly complex with lots of different variables. To make matters worse, each brain-scanning machine produces data stored in a different way or using different coordinate systems. This leaves researchers with the task of data transformations across systems, which is both time-consuming and error-prone.
“If everyone is doing their own pipeline, developed on their own, and we’re not cross-referencing them to one another, it makes it easier for bugs to find their way in,” Hansen explained.
Hansen and a team of researchers at The Montreal Neurological Institute-Hospital noticed these obstacles in their own work and decided to build a Python program to address them.
“When it comes to a lot of software toolboxes, it’s best to make one if you think it’s going to be useful to yourself,” Hansen said. “It was like, we have this problem, probably other people have this problem too.”
Since neuromaps is open-source and publicly accessible on GitHub, other researchers have been able to make use of it for their own projects. Others can see the code it employs and suggest changes based on bugs they encounter or use-cases they have found.
“[Users] would let us know, like, very specific bugs or things that we hadn’t caught, because it’s just […] not how we would have used it,” Hansen said. “So we were getting a better idea of how people were using it and that helped us develop it further. That’s definitely an ongoing process.”
According to Hansen, development and collaboration with other researchers, and even across disciplines, is critical to projects like neuromaps. In order to develop the software, they needed not just expertise in neuroscience, but also computer science, mathematics, and data management.
For the team behind neuromaps, the philosophy of collaboration and prioritizing multidisciplinary approaches goes beyond the development of the software itself.
“We’re definitely hoping that it’s going to help spark more multidisciplinary, cross-modal research. So that is to say, researchers being able to make bridges to many different disciplines, but also many different types of data,” Hansen said.
By allowing researchers to look across different types of data, even those collected by scientists in other branches of neuroscience, neuromaps allows researchers to connect with other labs and examine larger research trends.
“I think there’s a lot of value to looking at things very zoomed in, because that’s how we get the best image quality and depth. But then what neuromaps tries to do is also give us that zoomed-out perspective,” Hansen said. “So now let’s bring everything together and see, can we see these big-picture relationships?”